Featured Publication
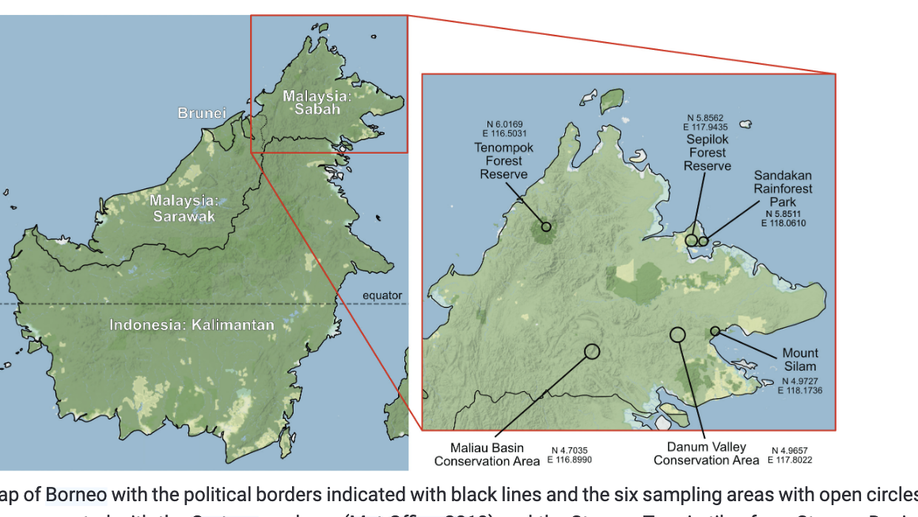
The Dacini fruit flies of Borneo: an annotated checklist with 89 species including three new to science (Tephritidae, Dacinae)
Our work adds 46 species to the 43 previously known from Borneo, bringing the total for the island to 89. Three new species are described: Bactrocera (Bactrocera) melanobivittata Doorenweerd, sp. nov., Dacus (Mellesis) danumensis Doorenweerd, sp. nov., and Zeugodacus (Zeugodacus) cataracta Doorenweerd, sp. nov.












